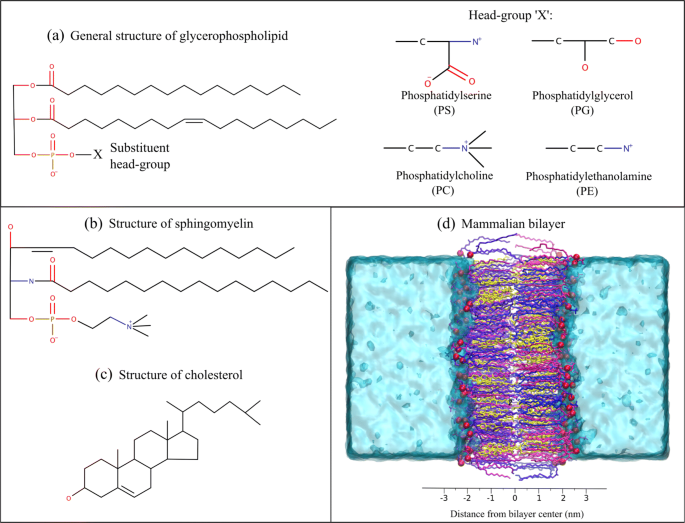
Molecular Dynamics Simulations of the Adenosine A2a Receptor in POPC and POPE Lipid Bilayers: Effects of Membrane on Protein Behavior | Journal of Chemical Information and Modeling

A coarse-grained approach to studying the interactions of the antimicrobial peptides aurein 1.2 and maculatin 1.1 with POPG/POPE lipid mixtures | SpringerLink

Palmitoyloleoylphosphatidylethanolamine (POPE) lipid bilayer, mo, Art Print | Barewalls Posters & Prints | bwc13426018

Multilayer Structures in Lipid Monolayer Films Containing Surfactant Protein C: Effects of Cholesterol and POPE: Biophysical Journal

Molecular structures of POPC a, POPE b, and POPG c with numbering of... | Download Scientific Diagram
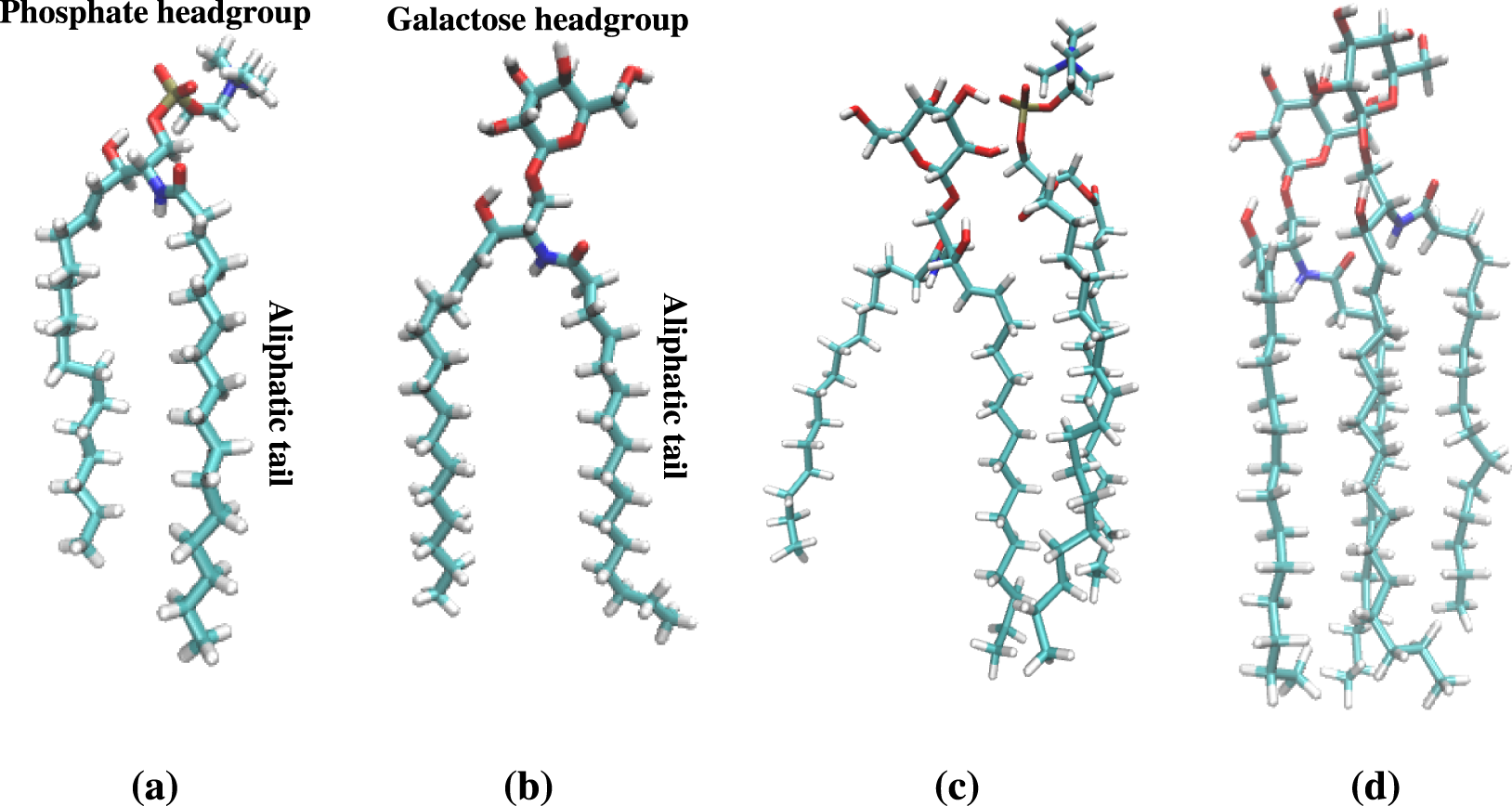
Role of lipid composition on the structural and mechanical features of axonal membranes: a molecular simulation study | Scientific Reports

Lipid shape is a key factor for membrane interactions of amphipathic helical peptides - ScienceDirect

IJMS | Free Full-Text | Residual Interactions of LL-37 with POPC and POPE:POPG Bilayer Model Studied by All-Atom Molecular Dynamics Simulation
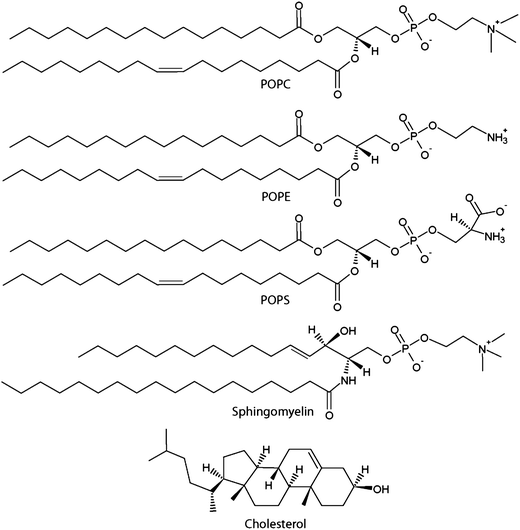
Biomimetic supported lipid bilayers with high cholesterol content formed by α-helical peptide -induced vesicle fusion - Journal of Materials Chemistry (RSC Publishing) DOI:10.1039/C2JM32016A

CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field. - Abstract - Europe PMC

Biomolecules | Free Full-Text | Examining the Effect of Charged Lipids on Mitochondrial Outer Membrane Dynamics Using Atomistic Simulations

Model parameters for simulation of physiological lipids - Hills - 2016 - Journal of Computational Chemistry - Wiley Online Library

Molecular structure with numbering of atoms and torsion angles of POPC... | Download Scientific Diagram
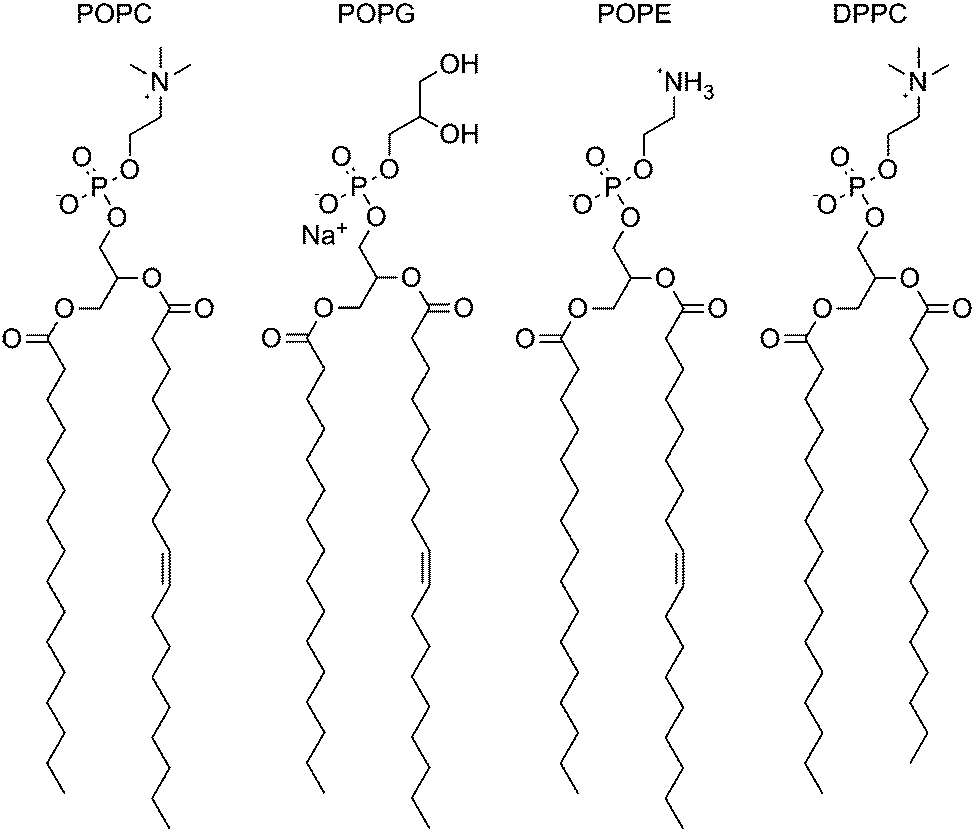
Anion transport across varying lipid membranes – the effect of lipophilicity - Chemical Communications (RSC Publishing) DOI:10.1039/C5CC00823A
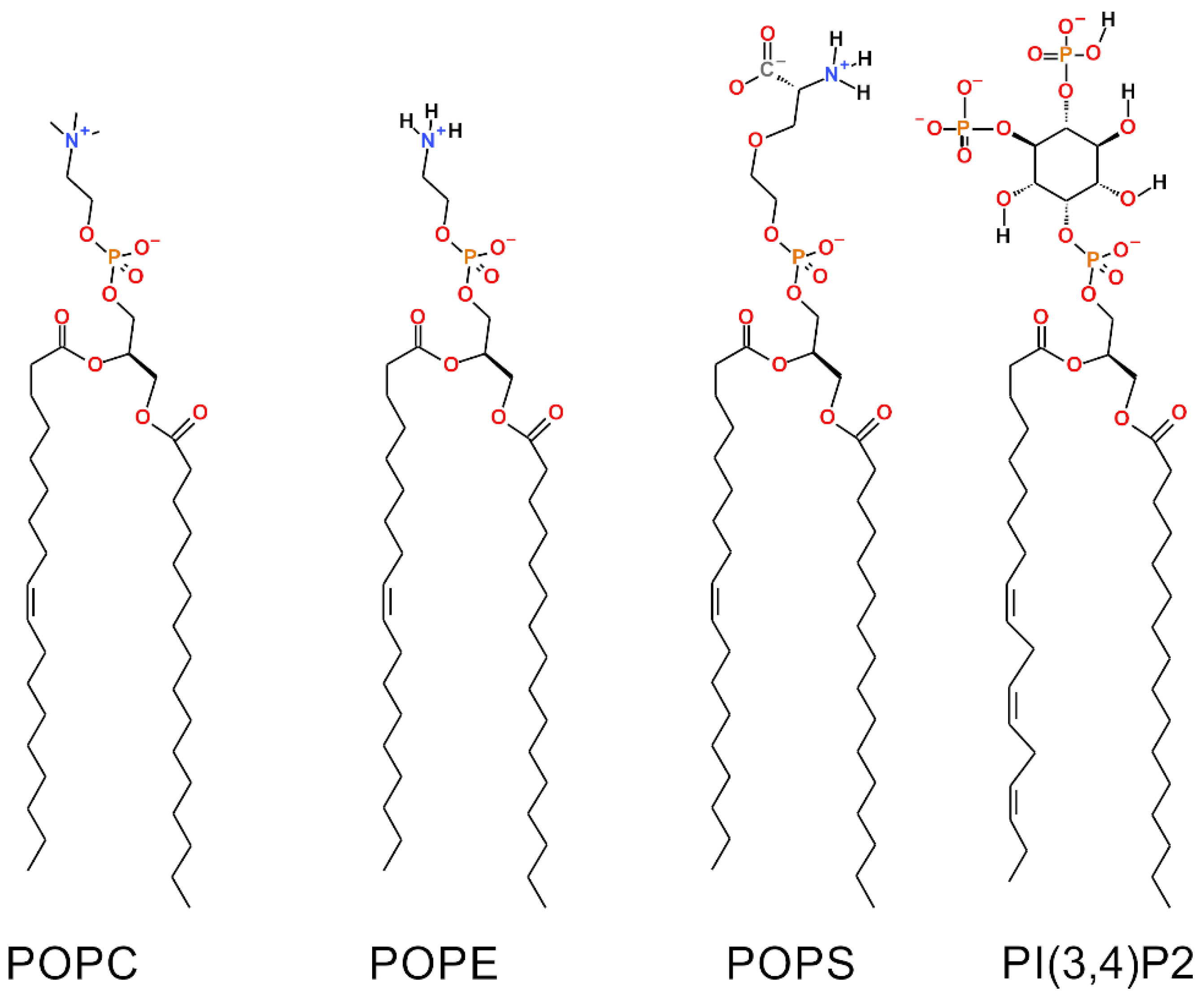
Biomolecules | Free Full-Text | Examining the Effect of Charged Lipids on Mitochondrial Outer Membrane Dynamics Using Atomistic Simulations
Interaction of POPC, DPPC, and POPE with the μ opioid receptor: A coarse-grained molecular dynamics study | PLOS ONE

Palmitoyloleoylphosphatidylethanolamine (POPE) lipid bilayer membrane in water, molecular model Stock Photo - Alamy
Interaction of POPC, DPPC, and POPE with the μ opioid receptor: A coarse-grained molecular dynamics study | PLOS ONE