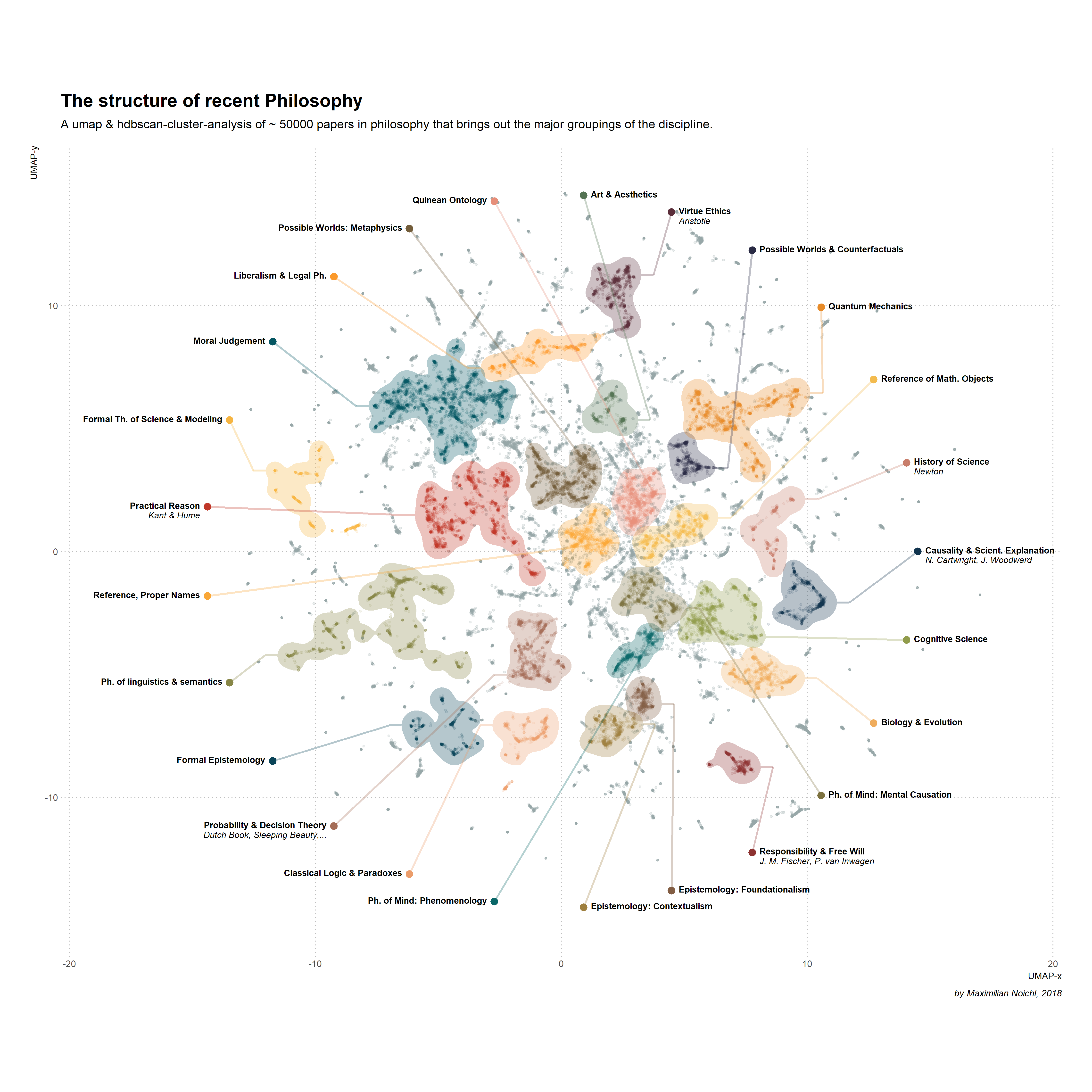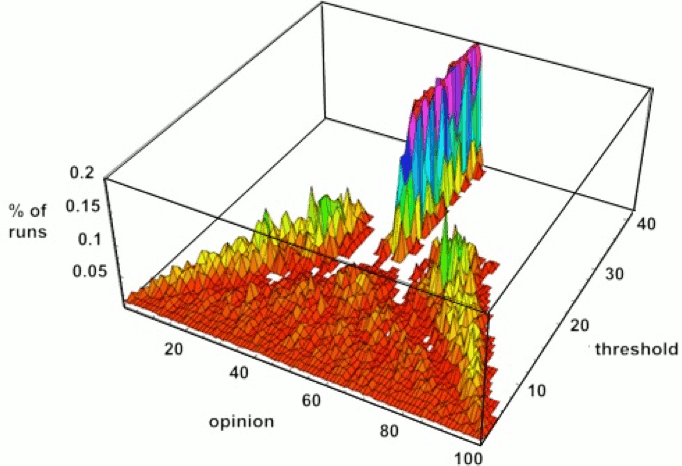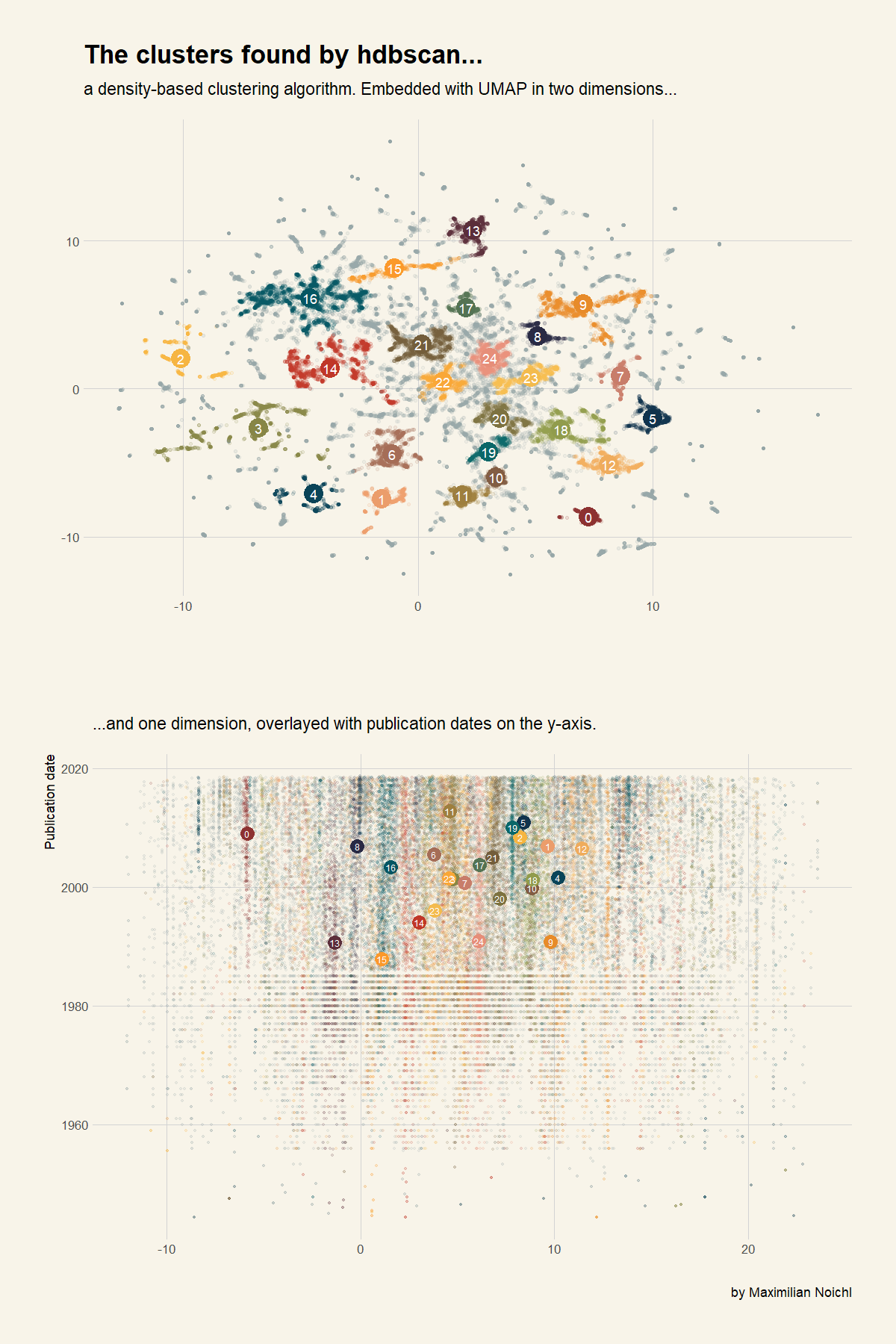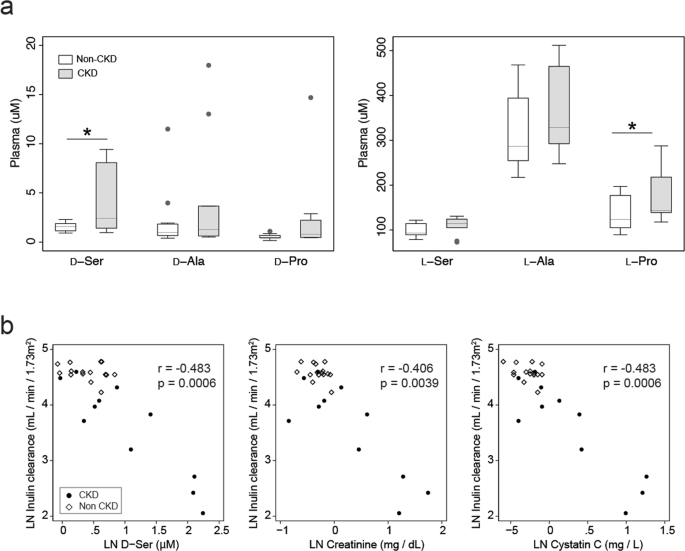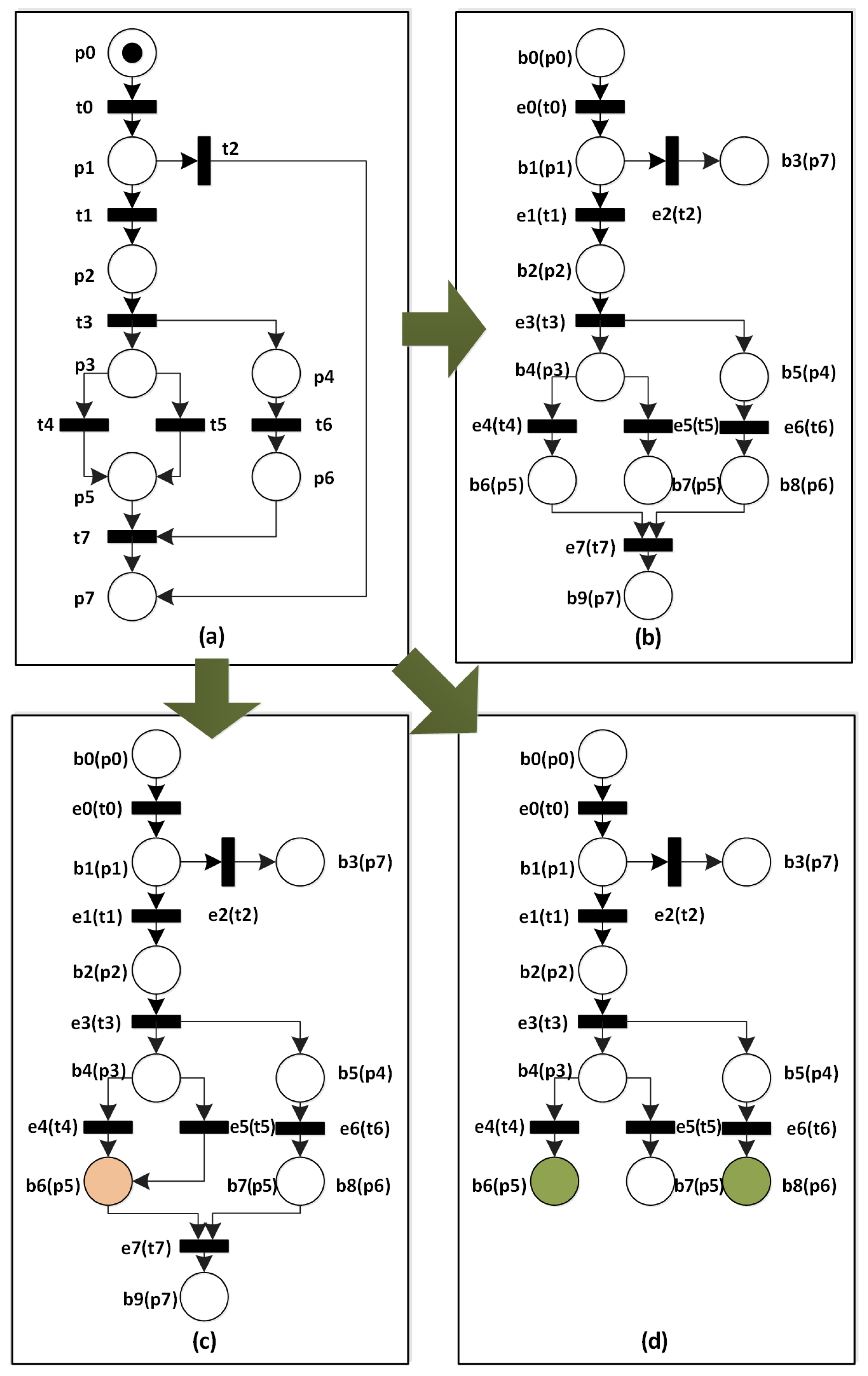
Symmetry | Free Full-Text | An Incremental and Backward-Conflict Guided Method for Unfolding Petri Nets | HTML

Cross-species neuroscience: closing the explanatory gap | Philosophical Transactions of the Royal Society B: Biological Sciences

Integrated transcriptional analysis unveils the dynamics of cellular differentiation in the developing mouse hippocampus | Scientific Reports
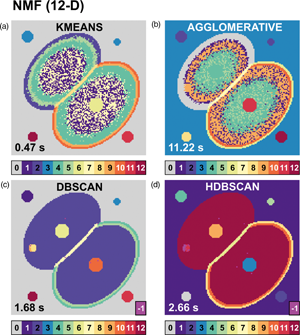
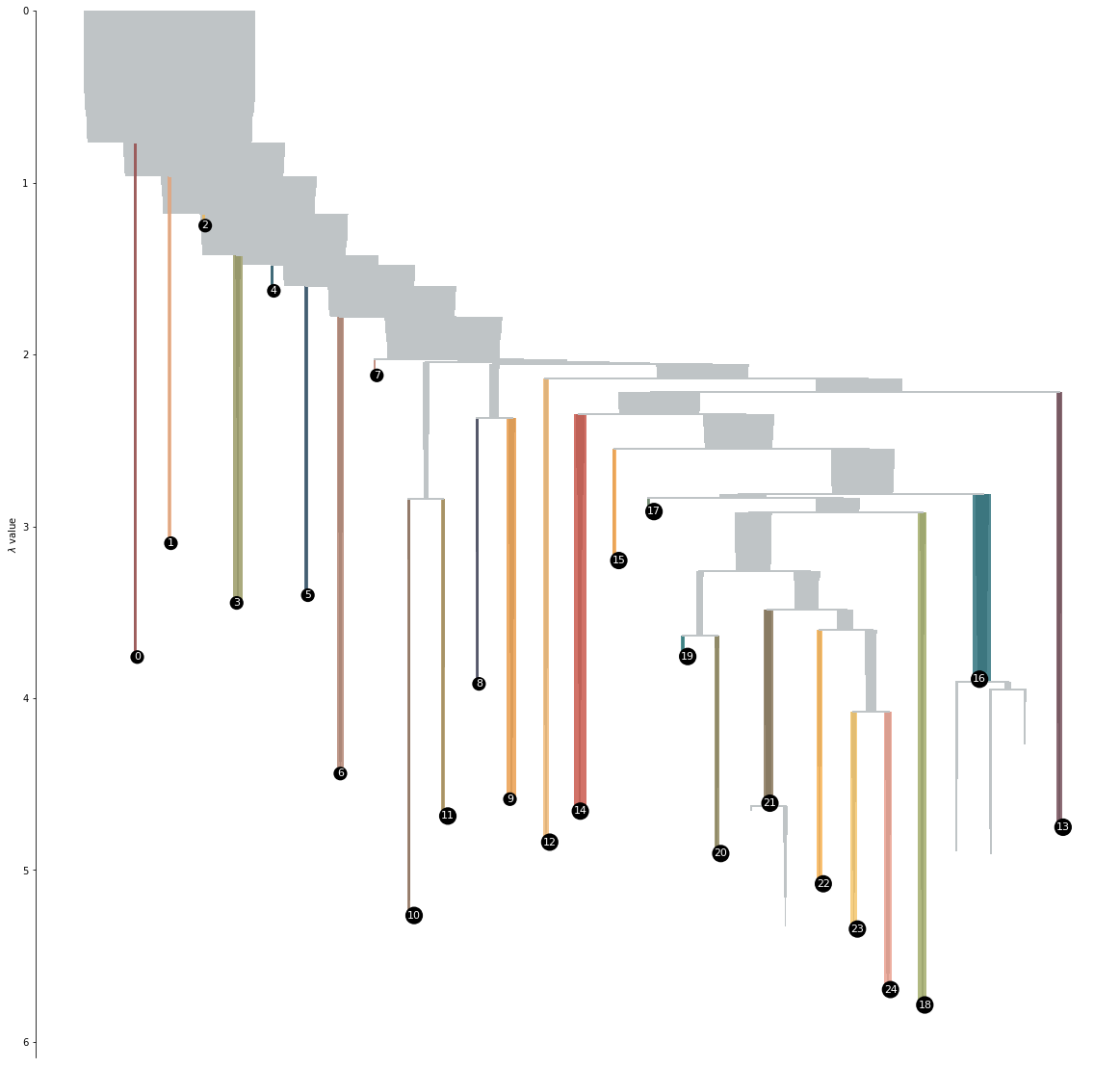
Strategies for EELS Data Analysis. Introducing UMAP and HDBSCAN for Dimensionality Reduction and Clustering | Microscopy and Microanalysis | Cambridge Core

Structural and functional auditory-related neuromarkers of dyslexia,... | Download Scientific Diagram

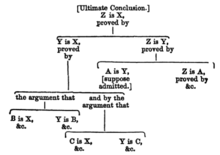
What Does 'Legal Obligation' Mean? - Wodak - 2018 - Pacific Philosophical Quarterly - Wiley Online Library
![Analysis of the genetic diversity and population structure of Salix psammophila based on phenotypic traits and simple sequence repeat markers [PeerJ] Analysis of the genetic diversity and population structure of Salix psammophila based on phenotypic traits and simple sequence repeat markers [PeerJ]](https://dfzljdn9uc3pi.cloudfront.net/2019/6419/1/fig-7-full.png)