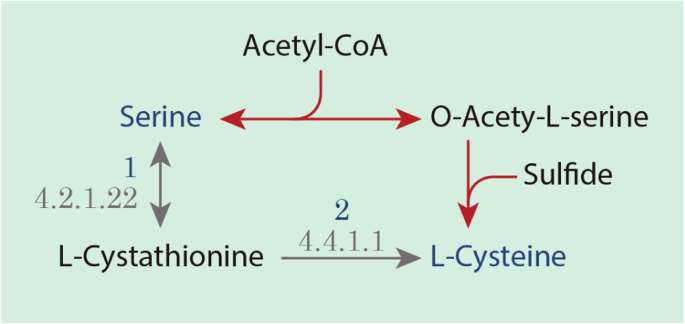
Genome-wide sequencing and metabolic annotation of Pythium irregulare CBS 494.86: understanding Eicosapentaenoic acid production | BMC Biotechnology | Full Text

Assessment of ligand binding site predictions in CASP10 - Gallo Cassarino - 2014 - Proteins: Structure, Function, and Bioinformatics - Wiley Online Library
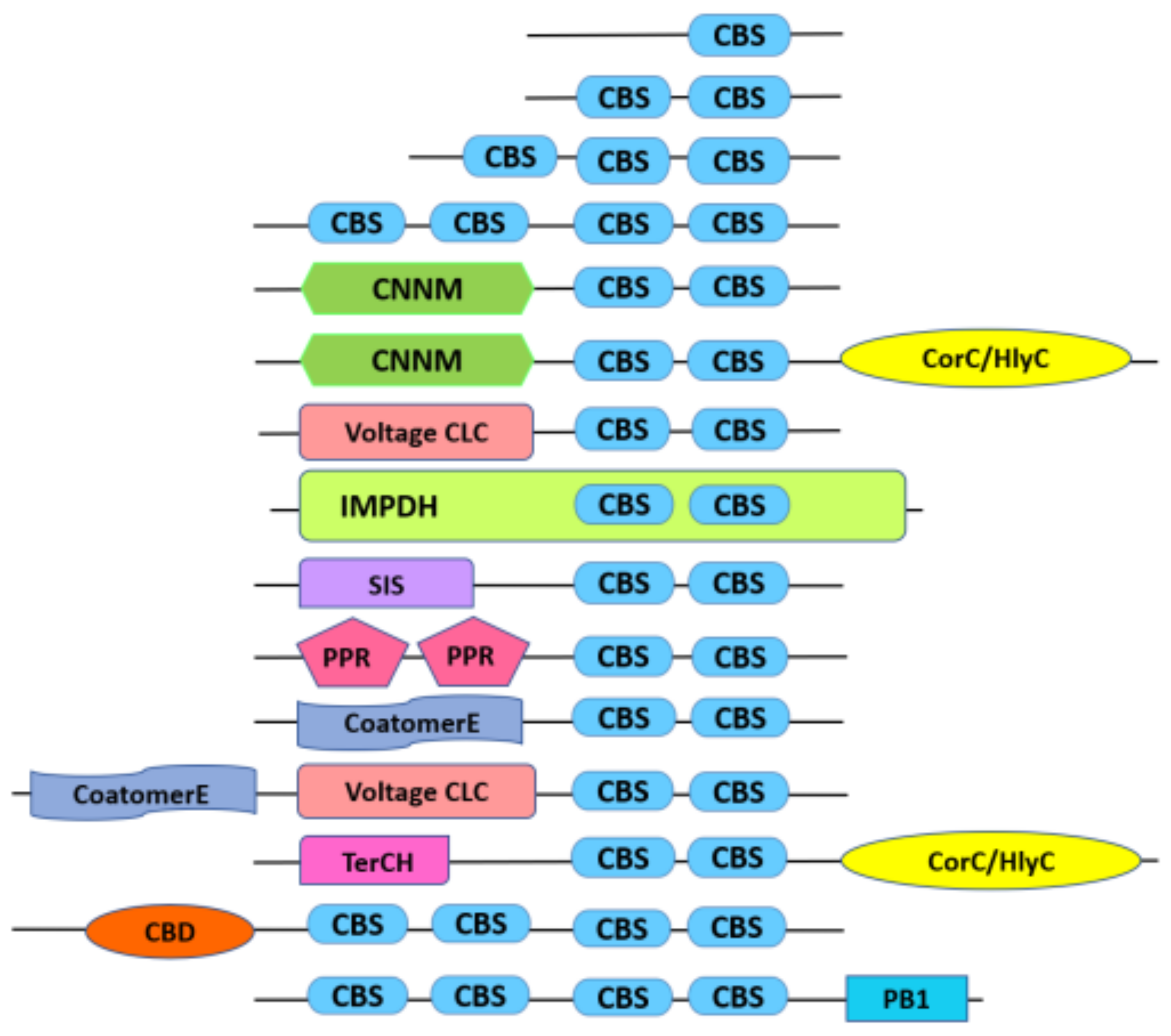
IJMS | Free Full-Text | Genetic Conservation of CBS Domain Containing Protein Family in Oryza Species and Their Association with Abiotic Stress Responses | HTML
![PDF] ScienceDirect Topology of membrane proteins — predictions , limitations and variations | Semantic Scholar PDF] ScienceDirect Topology of membrane proteins — predictions , limitations and variations | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/43668a1d2b7f3f251fc93da3b722ac5cd5a54241/6-Table1-1.png)
PDF] ScienceDirect Topology of membrane proteins — predictions , limitations and variations | Semantic Scholar

A Conserved Class II Type Thioester Domain-Containing Adhesin Is Required for Efficient Conjugation in Bacillus subtilis | mBio
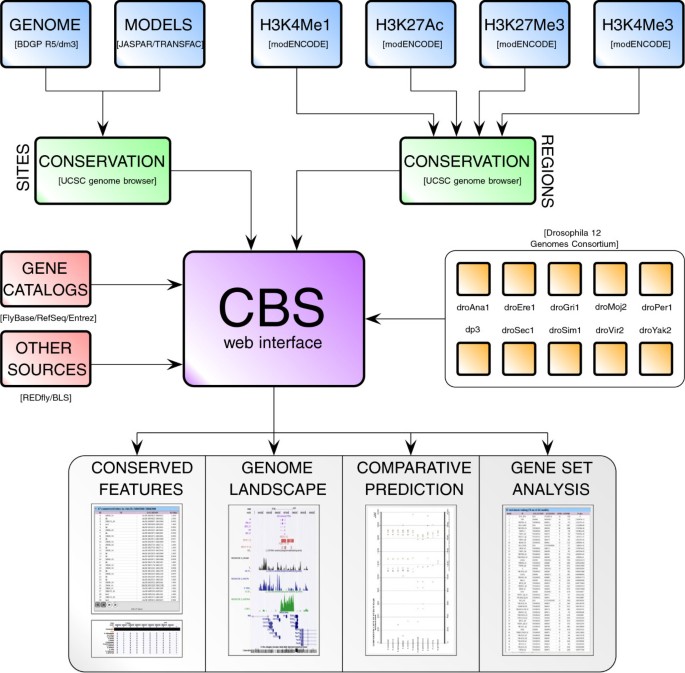
CBS: an open platform that integrates predictive methods and epigenetics information to characterize conserved regulatory features in multiple Drosophila genomes | BMC Genomics | Full Text
Predicting peptide MHC interactions Morten Nielsen, CBS, Department of Systems Biology, DTU. - ppt download
GitHub - isoprenaline/RTMHMM: Mouse Gene Only! Prediction of transmembrane helices in proteins by TMHMM Server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM ),a big Thanks to TMHMM! Input a csv file with Ensembl Gene IDs and
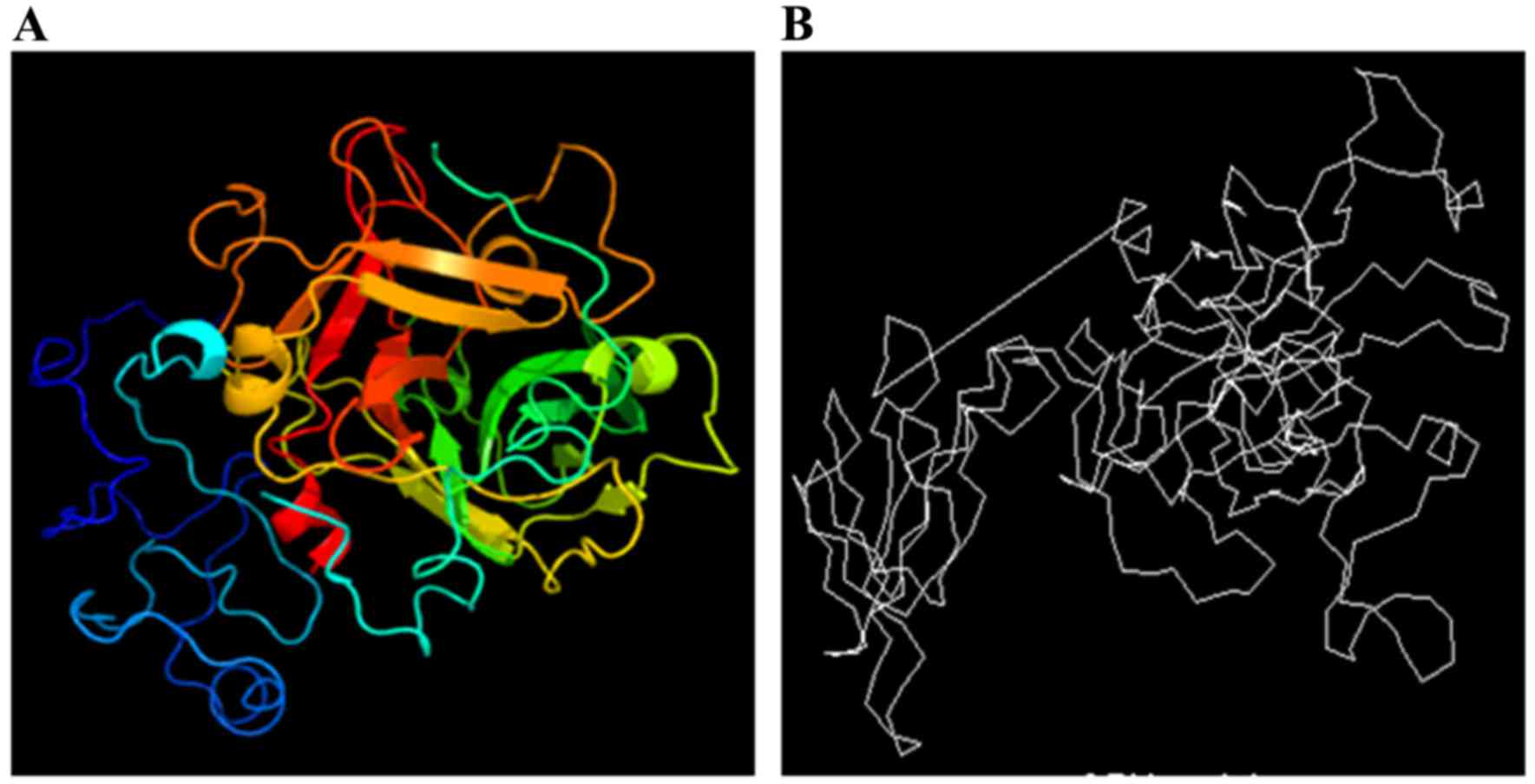
Center for Biologisk Sekvensanalyse Nikolaj Blom Center for Biological Sequence Analysis BioCentrum-DTU Technical University of Denmark - ppt download
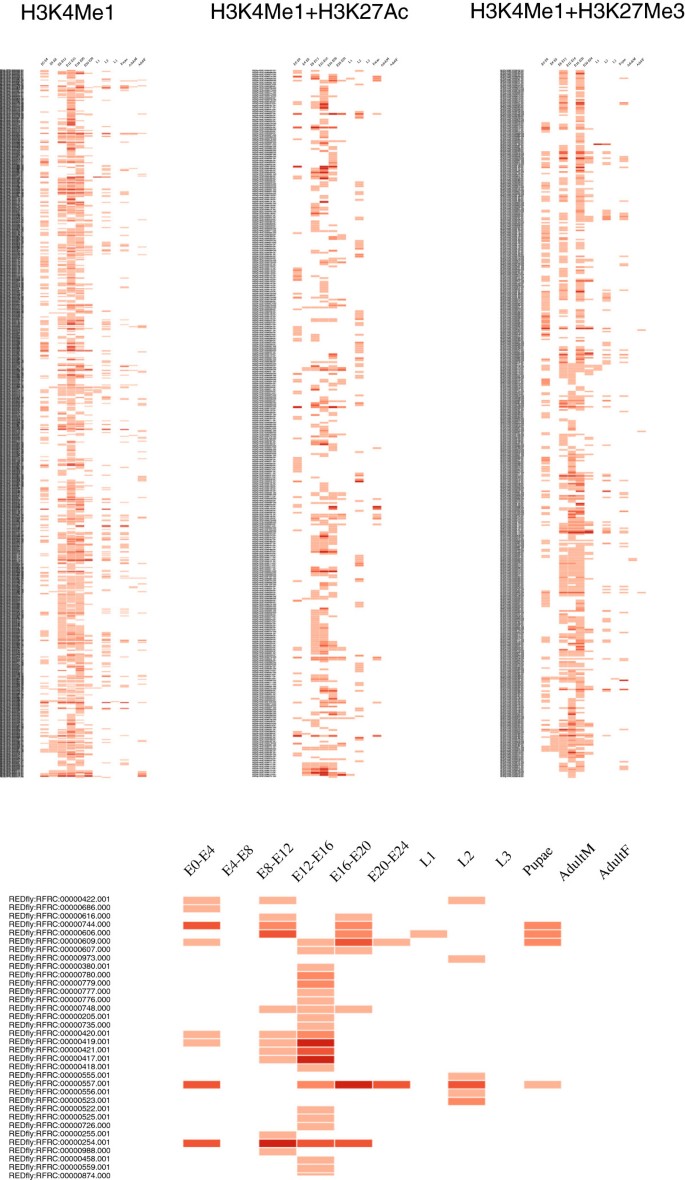
CBS: an open platform that integrates predictive methods and epigenetics information to characterize conserved regulatory features in multiple Drosophila genomes | BMC Genomics | Full Text